Science cluster

Summary
FAIRFUN4Biodiversity aims to enhance functional annotation of genomic resources generated by the biodiversity genomics community, particularly for non-model organisms. By leveraging AI-driven methodologies, this project seeks to generate publicly accessible functional data, promote Open Science practices, and improve cross-domain interoperability. The project will ensure compliance with FAIR principles, and will expand the portfolio of open access tools readily available for the biodiversity genomics community, ultimately contributing to a deeper understanding of the functional landscape of non-model organisms.
Challenge
Open Science project, Cross-domain/Cross-RI
Understanding the evolution of coding genes and their functions is crucial in evolutionary biology, yet many protein-coding genes remain poorly characterised, particularly in non-model organisms. This lack of functional annotation, especially in what is termed the 'dark proteome' - genes within a proteome without functional annotation - leads to incomplete models of evolutionary change and limits the identification of conserved or lineage-specific features. Traditional homology-based methods often fail to adequately transfer functional annotations. With the advent of initiatives such as the European Reference Genome Atlas or ATLASEA, where new genomes from non-model organisms are being sequenced and released daily, we need to leverage faster and scalable sequence-based functional prediction methods. In this line, embracing concepts from orthogonal disciplines, such as computer science and Artificial Intelligence (AI), could alleviate the problem.
Solution
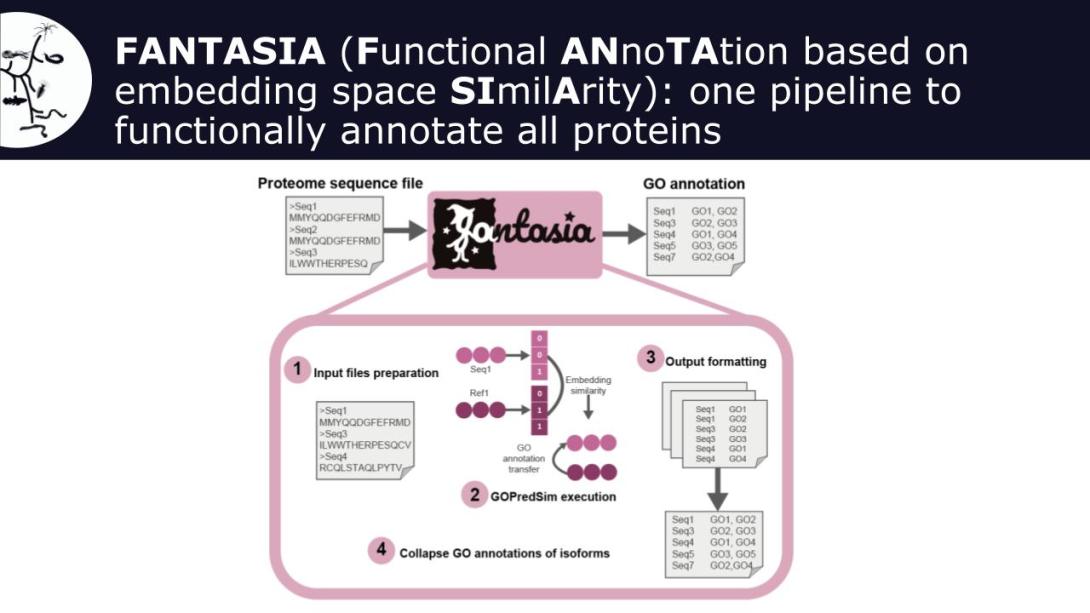
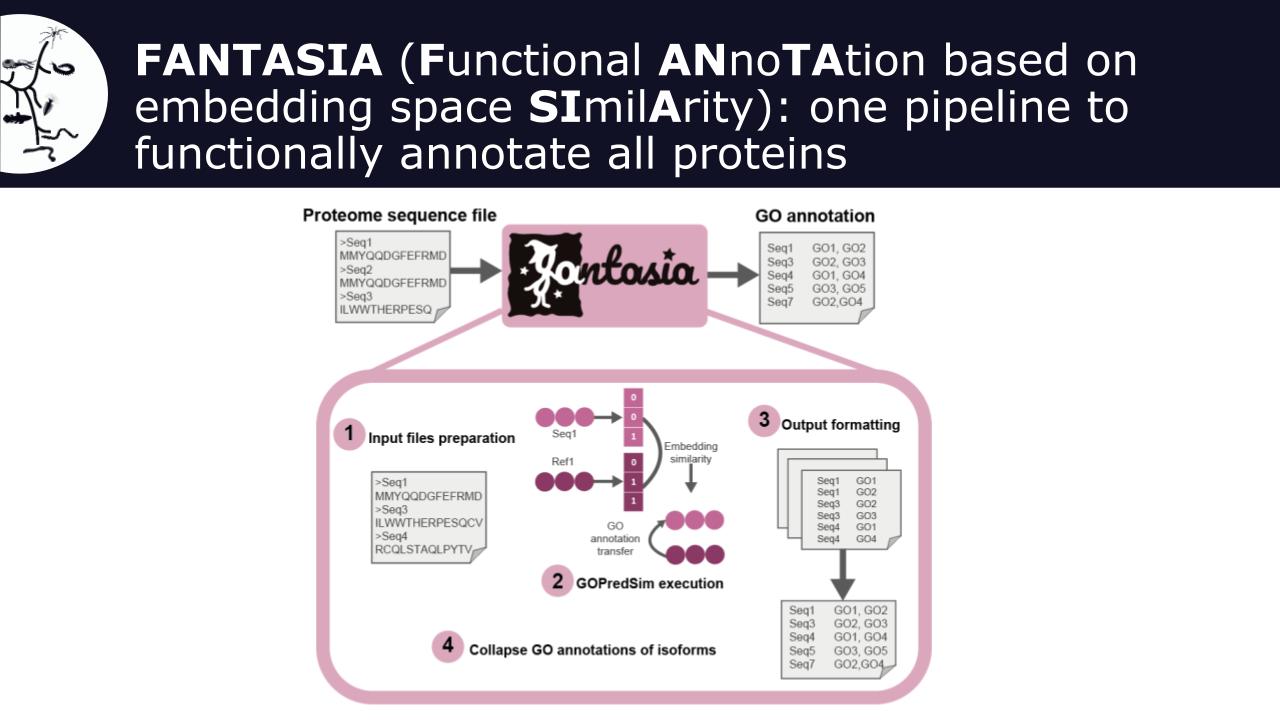
FAIRFUN4Biodiversity addresses these challenges with FANTASIA (Functional ANnotation based on embedding space SImilArity), a novel pipeline leveraging AI models from natural language processing, which overcomes the current limitations of homology-based methods and recovers functional annotation with great informativeness for virtually all genes in a proteome. The tool is currently available as an open access Singularity container.
To make it fully FAIR-compliant, the project will i) generate functional annotation data of the genomic resources generated by the biodiversity genomics community, and make all data publicly available to the research community; (ii) provide publicly-available functional annotation of genomes generated by the current biodiversity genomics consortia, such as the European Reference Genome Atlas (ERGA) or ATLASEA; (iii) Engage with the biodiversity genomics community for knowledge transfer and connect to RIs within the Science Clusters such as ELIXIR or LifeWatch; (iv) Improve the tool by leveraging bilingual models that take into account both natural language processing algorithms and protein structure inference ones, such as ProstT51.
Scientific Impact
This project significantly advances biodiversity genomics by addressing functional annotation for non-model organisms. By expanding publicly available functional annotation of the generated genomic resources, FAIRFUN4Biodiversity enhances downstream biological analyses and fosters collaboration among European RIs, such as ELIXIR and LifeWatch. Ultimately, it aims to deepen our understanding of the functional landscape of non-model organisms, stimulate Open Science practices in functional annotation of genomes of non-model organisms, improve cross-domain interoperability, and strengthen long-term coordination within the biodiversity genomics community at the European level.
Results
- FANTASIAV2 tool repo, registered in biotools
- FANTASIA has been recommended as annotation tool for coding proteins in the 1st version of the Report on Annotation Recommended Tools published in December 2024 by the Earth Biogenome project.
Publications
- Cases,I., Martínez-Redondo, GI, Fernández, R., Rojas, A.M.. Functional annotation of proteomes using protein language models: a high-throughput implementation of the ProtTrans model. In: KC, D.B. (eds) Large Language Models (LLMs) in Protein Bioinformatics. Methods in Molecular Biology, vol 2941. Humana, New York, NY. | DOI
- Vargas-Chávez, C., Benítez-Álvarez, L., Martínez-Redondo, G.I. et al. An episodic burst of massive genomic rearrangements and the origin of non-marine annelids. Nat Ecol Evol 9, 1263–1279 (2025) | DOI
Events
- 20 February, 2025 | Barcelona, Spain - Invited talk at the Barcelona Super Computer Center - Seminar about AI in functional annotation, presentation of FANTASIA
- 5-9 May, 2025 | Barcelona, Spain - Workshop on Genome Annotation
- 25 May, 2025 | Bordeaux, France - Invited talk at the CMF Unit in Bourdeaux - Seminar about AI in functional annotation, presentation of FANTASIA
- 20-24 July, 2025 | Liverpool, UK - ISMB2025 in the Comparative genomics and evolution track
Principal investigator

Rosa Fernández leverages phylogenomics and comparative genomics to understand how animals colonised land environments from marine ancestors (i.e., the origin of animal terrestrial biodiversity) and adaptation to life in caves. Ana Rojas investigates protein function and evolution and their relationship with structure /function through AI-based methods, in particular protein Language Models. Aureliano Bombarely is interested both in the development of bioinformatic tools and pipelines for genomic analysis, and in the study of how genomic information evolves associated to plant domestication and diversification using genomic tools.