Science cluster & challenges

Summary
The project Implementing FAIRness in structure-based drug design through Fragalysis Cloud aims to enhance the transparency and accessibility of structure-based drug design (SBDD) data by making it fully open and compliant with FAIR principles. Leveraging Diamond’s XChem service and the Fragalysis Cloud platform, the project will streamline data deposition and establish a first-of-its-kind tool for sharing, exploring, and evolving SBDD experiments. This will significantly lower the barrier to open data sharing, advancing drug discovery methodologies.
Challenge
Open Science project, Open Science Service, Cross-domain/Cross-RI
There is a strong consensus in the field of medicinal chemistry, that existing datasets for exploring and improving structure-based drug design (SBDD) are highly inadequate. The state-of-the-art of data availability has evolved into the separate repositories of the Protein Data Bank (accessed through PDBe in ELIXIR), and the ChEMBL database. These are individually well-developed and highly FAIR, but only for very specialised informaticians, and with necessarily strictly constrained ontologies. Moreover, data is often fragmented across various tools and hidden in unstructured formats, making it difficult for researchers to access and build upon existing datasets
Solution
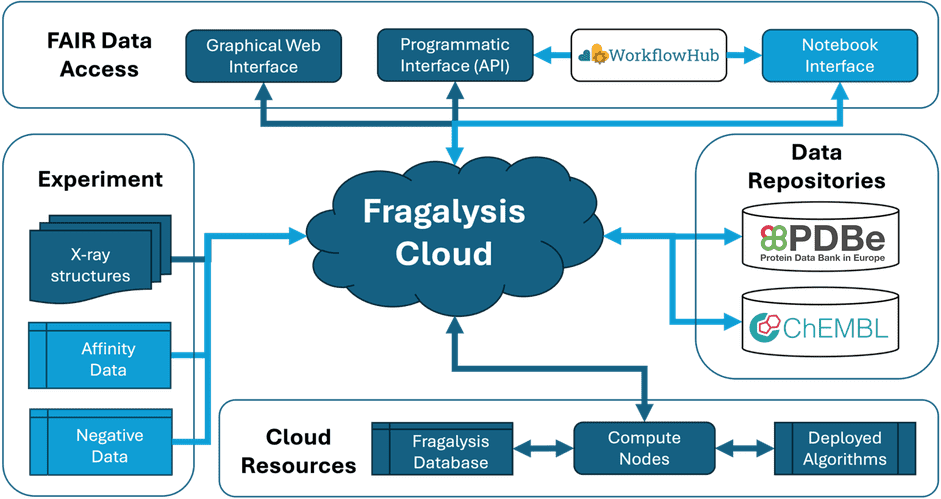
The project aims to enhance the Fragalysis Cloud platform to facilitate open and FAIR-compliant sharing of SBDD data. Fragalysis Cloud is a collaboration platform developed at Diamond’s XChem facility, used for curating, sharing and disseminating views of 3D data, and implementing best-practice medicinal chemistry algorithms and workflows for progressing results from fragment screens through early DMTA cycles. The project will map out clearly how Fragalysis can be a platform for collectively annotating and depositing data to PDBe and ChEMBL. Furthermore, it will make details of annotation workflows and computations accessible from those repositories, and generalised workflows will be made available to WorkflowHub.
By integrating data from 50–150 experiments, the project will create an accessible mechanism for depositing and querying large-scale datasets. Collaborating with key INSTRUCT (XChem) and ELIXIR services (PDBe and ChEMBL), which host the key data categories of the field (3D structures of protein-compound complexes and assay results of the compounds, including biochemical activity, biophysical affinity and ADME measurements), the project will enable researchers to explore SBDD projects with comprehensive data on context, provenance, design rationales and inspection of computational analyses.
Scientific Impact
The project will revolutionise the accessibility and reuse of SBDD data, providing an innovative tool for examining collective datasets in a FAIR manner. By making previously isolated data (and metadata) available for querying across projects, the project will improve transparency and reproducibility of experiments, while allowing to advance methodology and algorithms in the field. Researchers and developers will benefit from the insights gained from shared workflows, while contributing significantly to the FAIR data landscape in the life sciences.
The scientific relevance of the tools will be stress-tested by applying the new tools to real-world XChem data, both historic and new, and training the user community in their use, thereby greatly expanding the quantity and quality of SBDD data in the public domain.
Principal investigator

Warren Thompson is a Senior Computational Scientist at XChem, Diamond Light Source that works on integrating software engineering, chemistry, and prototyping to make open-source fragment-progression technologies, that include structural data dissemination, FAIR-data, collaboration tools and digitised chemistry, accessible to the fragment progression community.