Science cluster

Summary
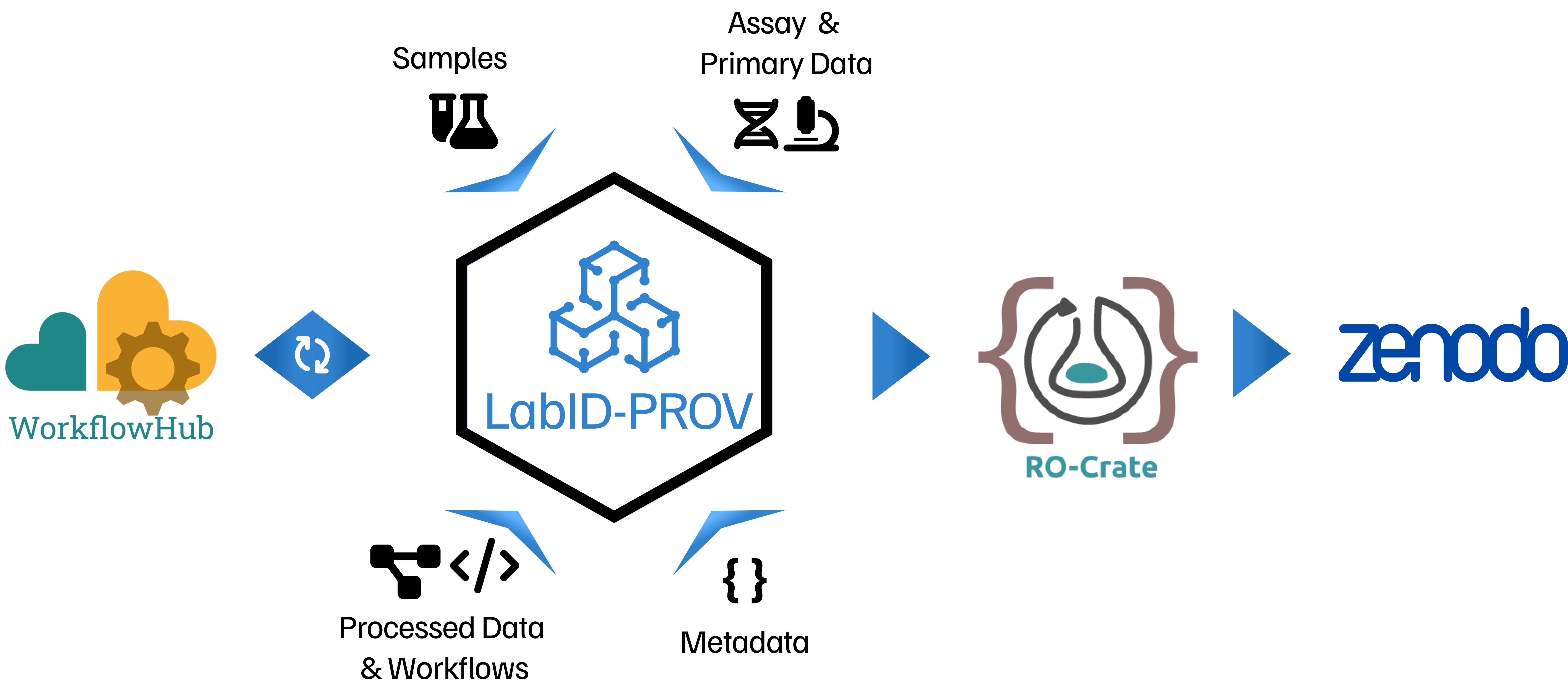
FAIR data management and the ability to track (meta)data from sample collection to the final computed (or derived) datasets have become increasingly crucial due to the increasing size and diversity of data generated across various scientific disciplines. However, to ensure reproducibility, comprehensive provenance metadata also needs to be available. Lab Integrated Data (LabID) is a web-based integrated platform designed to help individual scientists, research groups and core facilities to better manage, annotate and share their experiments, assays, samples and datasets actively, in compliance with FAIR principles. While in LabID processed data can already be stored and connected to its primary data, associated assays and original samples, accurate modelling of both workflow (WF) and WF runs is currently lacking. The LabID-PROV project aims to extend the LabID data model to include these concepts, offering a unified application to manage derived data provenance independently from analysis procedure and platform, and providing a concrete solution to ensure the traceability of derived data.
Challenge
Open Science Service, Main RI concerned, Cross-domain/Cross-RI
Managing the increasing volume and variety of (meta)data - from sample collection to derived datasets - poses significant challenges in scientific research. While primary data is regularly shared in repositories, the sharing of final derived datasets remains inadequate. This is often due to the lack of comprehensive provenance metadata that ensures reproducibility and trustworthiness. Furthermore, the diverse languages, tools, and computing environments used by various stakeholders complicate the tracking of workflows and their corresponding metadata, limiting the reusability of shared data and hindering Open Science efforts.
Solution
LabID-PROV proposes to enhance the LabID platform by allowing accurate modelling of both WFs and WF runs. To this end, the project will use several resources in the LS RI Science Cluster, namely, WorkflowHub, a resource for WF indexing, discovery and re-use; RO-Crate, which can be used to package and share research objects including WF and WF runs using the WF and WF Run RO-Crate profiles; Galaxy, a web-based analysis platform; and Zenodo, an open platform for preserving and sharing research output. The main goal is to streamline the import of datasets (and their metadata) described using Workflow Run RO-Crate profiles into LabID. Also, the project will implement use cases using both omics and imaging data, reflecting real-world scenarios, to demonstrate the integration of the new LabID capabilities into the existing Open Science landscape. These use cases will be used to generate online LabID tutorials demonstrating best practices in WF development and the FAIR dissemination of associated derived data with their provenance. Finally, wider dissemination of this work will be achieved by registering the training in the Training eSupport System and through a future LabIDPROV workshop.
Scientific Impact
The LabID-PROV project aims to bridge critical gaps in data traceability to facilitate FAIR sharing of derived data together with their provenance metadata, through the integration of LS RI Science Cluster tools and standards. LabID-PROV not only will enhance the reproducibility of scientific findings but also encourages researchers across various disciplines to adopt robust data management practices. The project will ultimately support a wider community of scientists, enabling them to efficiently share valuable research outputs and contribute to a more sustainable Open Science ecosystem.
Results
- Release of LabID 25.2.0: Version 25.2.0 of LabID was the first to include features for workflow management. Among the new functionalities are the possibility to define workflows and document separate versions of a workflow. Workflow can be imported from Git repositories or other dedicated repositories like Galaxy and WorkflowHub. These new features are the first steps towards documenting provenance for data produced by computational workflows. One of LabID core features is to document the provenance of data, from biological samples to the raw data produced by a specific instrument and experimental protocol. The workflow integration will enable extending this provenance principle to data derived from computational workflows used to analyze or process the raw data. LabID will thus ensure end-to-end data provenance, while fostering novel FAIR practices for the documentation of derived datasets.
- Release of LabID 25.8.0: Version 25.8.0 of LabID finalized the workflow management functionalities by providing support for documentation of computational workflows execution, so called Workflow Runs. A workflow run has a reference to the workflow version, the (raw) data used as input, and the “derived data” generated by the run. By associating this information, LabID ensures data-provenance is captured end-to-end: from biological sample, to raw data coming out of an instrument, and finally derived data produced when processing the raw data with computational workflows. Command line utilities were also developed as part of the release, to facilitate the registration of workflow runs in batch. Besides workflows and workflow runs can be exported as RO-Crate, for instance for publication on WorkflowHub.
LabID is listed on the official Research Object Crate website among the list of software using the RO-Crate specification. The page briefly describes how LabID takes advantage of the Workflow and Workflow Run RO-Crate specification to import/export workflows and workflow runs, for instance from WorkflowHub.
The team contributed to the official ro-crate-py python library that can be used to parse and generate RO-Crates. The contributions include the addition of a new function to facilitate the creation of Workflow RO-Crate (https://github.com/ResearchObject/ro-crate-py/pull/221) and “subcrates” (https://github.com/ResearchObject/ro-crate-py/pull/244), the addition of an automatic code formatting routine (pre-commit-hooks see https://github.com/ResearchObject/ro-crate-py/pull/245) and some bug reports (https://github.com/ResearchObject/ro-crate-py/issues/222).
Publications
- TRaversing European Coastlines (TREC) Live Plankton High-Content Fluorescence Microscopy Dataset, DOI - This published dataset made use of LabID for data-management and metadata curation (see acknowledgments)
Events
- 1-3 October 2025 | Freiburg, Germany - European Galaxy Days. This yearly event brings together researchers using the Galaxy workflow management system, developers of the platform and maintainers of Galaxy server instances. Charles Girardot from the team attended the event, presenting a poster describing the LabID-PROV OSCARS project, and how Workflow and WorkflowRun can be imported from Galaxy | Poster.
- 14-16 October 2025 | EMBL-EBI, Hinxton, UK - EMBL data-science week. Charles Girardot presented LabID for data management and provenance at the annual EMBL data-science week.
- 1-5 December 2025 | Walsrode, Germany - de.NBI Hackathon. Laurent Thomas attended the hackathon to work on improving software tools to handle Research Object Crates (RO-Crates) in the context of distributed storage environments. The hackathon resulted in a proposal to support “referencing” subcrates within a top-level crate. This is discussed in the following pull-request on GitHub. Pictures
- 28-30 January 2026 | EMBL Heidelberg, Germany - 3C-CoDash Hackathon. As part of this hackathon, Laurent Thomas co-organises a project around FAIR image data-management. The session will illustrate the usage of LabID to track how images are converted to OME-Zarr as a workflow run, thus ensuring the provenance of the converted data is documented.
- 24-27 March 2026 | Vienna, Austria - Poster presentation at FAIR DO Conference (TBC)
Promotional material
- POSTER | LabID workflow integration: recording data-provenance from sample to workflow results (presented at the European Galaxy Days by Charles Girardot)
Principal investigator

Eileen Furlong is head of the Genome Biology dept and Senior Scientist at EMBL. She studied biochemistry at Univ. College Dublin, where she obtained her Ph.D. and moved to developmental biology for her postdoc at Stanford Univ. She became a group leader at EMBL in 2002, and is head of the Genome Biology Dept. since 2009. She is a recipient of two ERC advanced grants, an elected member of EMBO, Academia Europaea, Leopoldina and Fellow of the Royal Society (FRS). Her work uncovered different mechanisms of genome regulation, including how enhancers function and regulate developmental programmes.
- LabID-PROV Infographics
- The open-source code repositories of LabID can be found on GitLab
- Release notes are available as part of the user-documentation.

