Science clusters

Summary
Computational models of multicellular biological systems are essential for understanding how tissues grow, maintain homeostasis, and transition into disease states, such as cancer. These mechanistic models capture interacting biological processes across cellular, tissue, and organ scales, enabling quantitative predictions. The MultiCellML-TDR project aims to facilitate FAIR computational models and Trustworthy Digital Repositories (TDRs) by advancing their standardisation under the ELIXIR-FAIRDOM and COMBINE umbrellas.
As a proof of concept using the advanced standardisation, the project will compose, execute and analyse larger computational models of the human liver and its diseases from submodels derived from at least three model repositories.
Challenge
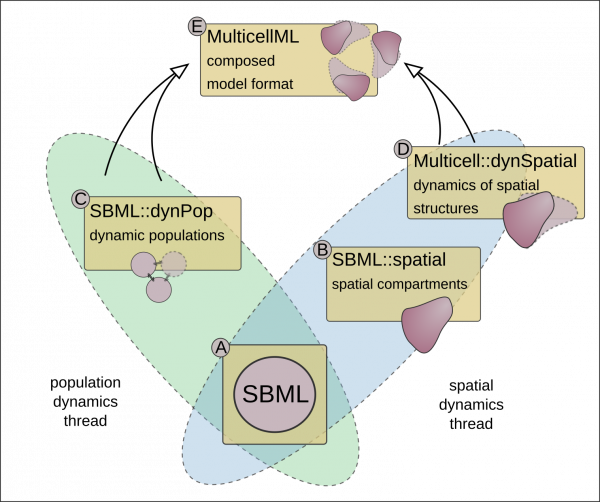
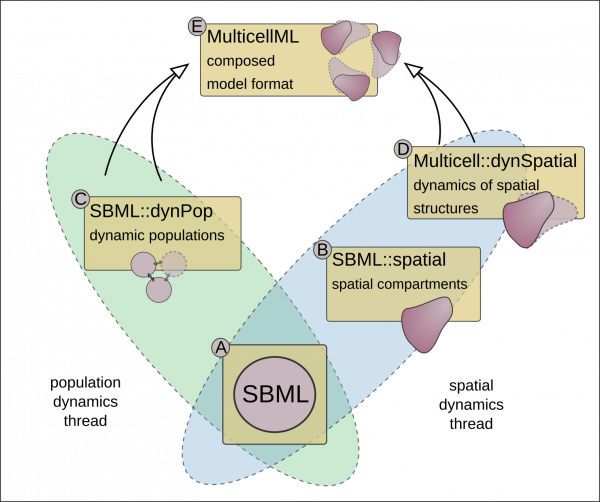
Computational models of complex multicellular biological systems often combine diverse mathematical formalisms such as partial differential equations, cell mechanics models, and agent-based models of dynamic cell behaviour. Unlike computational models for biochemical processes - supported by established standards and Trustworthy Digital Repositories (TDRs) such as ELIXIR’s BioModels - multicellular models are just starting to gain support by standards. Addressing this gap is essential for closer collaboration and modular extensibility of models, both critical to biomedical research and Virtual Human Twins.
Solution
MultiCellML-TDR will advance the standardisation of computational model descriptions for multiscale and multicellular biological systems using a modular composition of multiple ELIXIR-FAIRsharing standards. It will extend existing model repositories for multicellular models, and transparently document the standardisation workflow. Newly developed software and tools will be registered through ELIXIR-bio.tools, while ELIXIR-Identifiers will be used to advance multicellular computational models’ annotation. Training activities through ELIXIR-TeSS will support users. These deliverables together will enhance FAIRness of computational models for multicellular biological systems. The project will showcase its approach by composing, executing and analysing integrated liver models sourced from ELIXIR’s BioModels repository, FAIRDOMHub and the domain-specific MorpheusML model repository.
Scientific Impact
Researchers and developers across the life sciences and beyond will benefit from the project’s modular standardisation approach, shared workflows and computational models. Its application to multiscale liver modelling - capturing collective cell behaviour driven by molecular, cellular and tissue-level processes - highlights how integrated models reveal emergent phenomena not accessible through single-scale approaches. Native compatibility of 3D tissue models with microscopy image data will enable a close collaboration with experimentalists.
Principal investigator

Lutz Brusch obtained a Diploma and a PhD in Physics for work on physico-chemical pattern formation at the Max Planck Institute for the Physics of Complex Systems in Dresden, Germany. As a postdoc, he focused on biological pattern formation and biophysics at the Centre de Bioingénierie Gilbert Durand in Toulouse, France, and the Riken Omics Science Center in Yokohama, Japan. He is a permanent research group leader at the high-performance computing center of TU Dresden developing user-friendly modeling and simulation tools. In close collaboration with experimentalists, his group is studying dynamical processes during embryo development and disease progression.