This short article is one in a series addressing the impact of the OSCARS project on Open Science practices in Europe. (Reading suggestion: start with the introduction to the series).
Read the article on Zenodo
Data annotation is the backbone of Open Science, because it transforms datasets from opaque files into well-described, discoverable, and reusable research assets. Data annotation is also the first way to increase data value, data linking, interoperability and reproducibility of research.
It is the process of labelling, or enriching data with structured metadata - tags, categories, comments, semantic links, or controlled vocabulary terms - that makes the dataset intelligible to both humans and machines. In fact, semantic annotation with controlled vocabularies (i.e., validated by the research community) allows machines to parse, discover, and integrate datasets at scale in the FAIR web of data, which is one of the main goals of Open Science also prioritised by the European Open Science Cloud (EOSC). Metadata are a key enabler for adherence to FAIR principles.
Annotations themselves can become FAIR digital objects. When datasets are enriched with sufficient, appropriate, and standardised annotations, they can be discovered and effectively reused in different fields, which is crucial to EOSC’s goal of promoting cross-disciplinary access to data and tools. To support this process, categorisation systems, catalogues, data dictionaries, and other data descriptors act as a backbone for data reusability across and between research communities.
There are, however, still important challenges to be addressed in the research community before data annotation becomes a common practice. Firstly, specific fields require tailored standards. Moreover, researchers require easy-to-use and adaptable annotation tools and support to generate accurate FAIR metadata efficiently. In this respect, scalable tools (e.g. AI‐assisted labelling) are being explored. Ensuring the quality and consistency of annotations remains essential.
To incentivise the research community to take up Open Science, EOSC foresees rewarding data annotation practices, embedding FAIR-aligned workflows at institutional and national levels as stated in the Strategic Research and Innovation Agenda - SRIA.
The SRIA embeds annotation deeply in EOSC: through interoperability guidance, FAIR-oriented core services, semantic standardisation, and alignment with data spaces. To realise the full potential, the ecosystem needs robust tools, continuously enriched domain specific vocabularies, and disciplined annotation practice enhanced by data stewardship support and assistance.
How have the Science Clusters addressed data annotation and the associated challenges?
All five Science Clusters support data annotation through complementary strategies - developing community-specific metadata schemas, supporting annotation tools and workflows, and embedding annotation practices in training and FAIR implementation.
An outline of the key contributions from each of the Science Clusters to the area of data annotation is provided below.
ENVRI (Environmental Sciences)
To enhance (meta) data annotation, the ENVRI Science Cluster promotes the use of a common metadata schema based on DCAT Application profile for data portals in Europe - DCAT-AP, facilitating machine-actionable annotations across environmental subdomains.
In addition, the FAIR Implementation Profiles (FIP) wizard [1], co-developed with GO FAIR, provides a list of declared technology choices, also referred to as FAIR Enabling Resources, that are intended to implement one or more of the FAIR Guiding Principles, made as a collective decision by the members of a particular community of practice.
Moreover, science demonstrators showing the capabilities of service provision among ENVRI Research Infrastructures and Science Clusters are built with Jupyter Notebooks in the Galaxy environment - an open-source web platform that allows one to create and share documents that contain live code, equations, visualisations, and narrative text.
ESCAPE (Astronomy, Nuclear and Particle Physics)
The ESCAPE Science Cluster fosters data annotation through its Data Lake infrastructure, supporting (meta)data-driven research data lifecycle management, whereas cross-border software and analysis platforms are enhanced with annotated software registries, supporting rich metadata and reuse. Virtual Research Environments (VREs) are also being developed with annotated services and workflows, designed to scale across RIs and Science Clusters.
LS-RI (Life Sciences)
The LS-RI Science Cluster has developed many domain-specific metadata schemas and minimum metadata requirements that serve as annotated descriptors of datasets, crucial for compliance, reproducibility, and reusability.
For instance, to enable the development and FAIR management of interdisciplinary workflows, LS-RI has established services such as WorkflowHub, RO-Crate, and Galaxy, which help structure and annotate data workflows, enabling traceability and reproducibility. Raw data can be processed with tools and workflows shared via WorkflowHub. Through their execution in Galaxy, the initial data are enriched with metadata, domain knowledge, and detailed process information, ensuring that each analytical step is transparent and reproducible. The resulting enriched outputs can then be packaged as FAIR digital objects, for example in the RO-Crate format, and exported into public archives, thus facilitating long-term accessibility, interoperability, and reuse.
To foster uptakes and reuse, annotation is embedded in community co-creation practices, such as hackathons, and FAIR training events that produce annotated datasets and workflows and harmonise them through crosswalks.
Moving forward, these practices call for governance models capable of validating commonly adopted annotation systems — from categorisation schemes that enhance findability, to sensitivity classifications that regulate access and protection, data typologies that enable interoperation and aggregation, and quality descriptors that underpin efficient mobilisation and data reuse at broader levels.
PaNOSC (Photon and Neutron Science)
The PaNOSC Science Cluster has been using and promoting the NeXus metadata standard and the PaNET vocabulary to provide semantic annotation of experimental data.
Moreover, through its federated search API, annotated metadata enables discoverability across photon and neutron experiments, whereas to improve access and traceability, PaNOSC has been exploring collaboration with SSHOC for Digital Object Identifier - DOI) annotation strategies.
PaNOSC has planned to also create new collaborations with scientific communities and publishers to promote the use of standard metadata and automatic verification before publication to ensure FAIRness, and thereby encouraging the increase in the quality of metadata and thus enhancing the findability of the data and supporting its reuse.
SSHOC (Social Sciences and Humanities)
The SSH Open Science Cluster (SSHOC) addresses challenges in heterogeneous annotation practices inherent to the disciplinary diversity within the SSH domain that features multilingual and semantically rich metadata. SSHOC focuses heavily on vocabulary alignment and metadata translations to enable interoperability of SSH data and resources from other domains.
The SSH Open Marketplace includes curated, annotated workflows, datasets, and tools, aligning them with EOSC services. Examples of SSHOC services for data annotation are the Data Management Expert Guide (DMEG), which provides annotation guidance to diverse communities, and the Language Resource Switchboard, which helps you find tools that can process your data.
OSCARS' contribution to better data annotation practices
OSCARS enhances cross-fertilisation between the Science Clusters by establishing common approaches to FAIR data management and data annotation. Concrete use cases and science demonstrators stemming from the OSCARS funded projects will help to adopt more FAIR data practices to support reproducibility and robustness in Open Science.
OSCARS is also committed to providing open access to OSCARS publications, code and associated data, including early access to and open annotation of OSCARS’ research results and deliverables, together with categorisation systems that permit indexation in both general and thematic catalogues (e.g. FAIRsharing).
Beyond project outputs, OSCARS contributes transversal tools that strengthen data annotation practices. Competence Centres must serve as a key instrument for mentoring and guiding communities in annotation workflows. Categorisation systems for data retrieval, sensitivity classification, quality assessment, and typologies are essential to improve interoperability and reuse. The forthcoming Data Steward Registry aims to provide a structured mechanism to identify available expertise, mentoring capacity, and reviewer willingness, with OSCARS project participants expected to be among the first to be listed. Complementing this, the Terminology Task Force — promoting the use of EOSC-recommended vocabularies and offering guidance on descriptors — constitutes another key OSCARS enabler for the widespread uptake of data annotation practices.
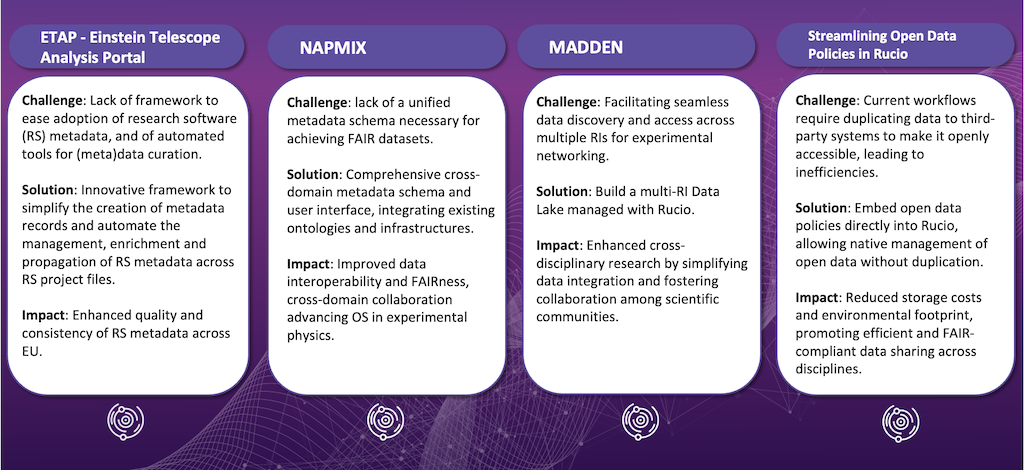
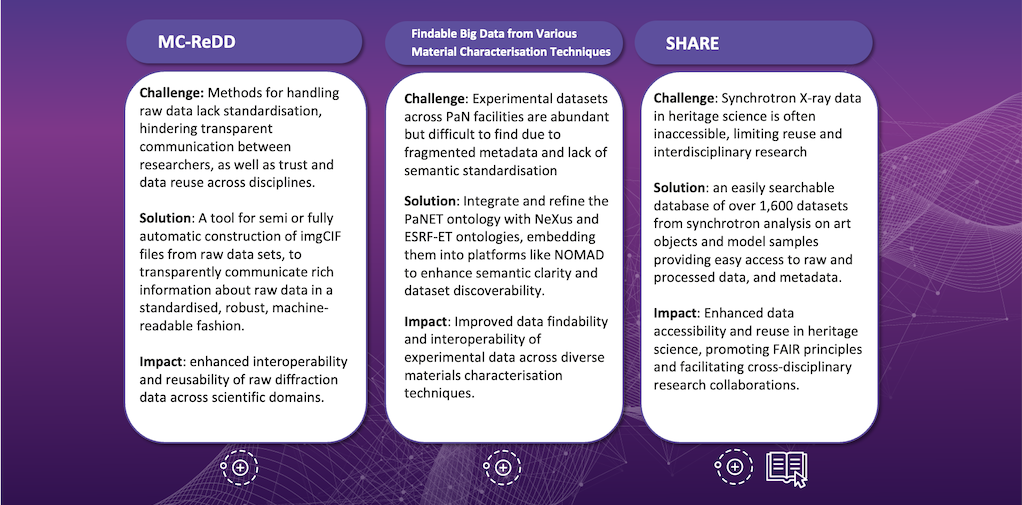
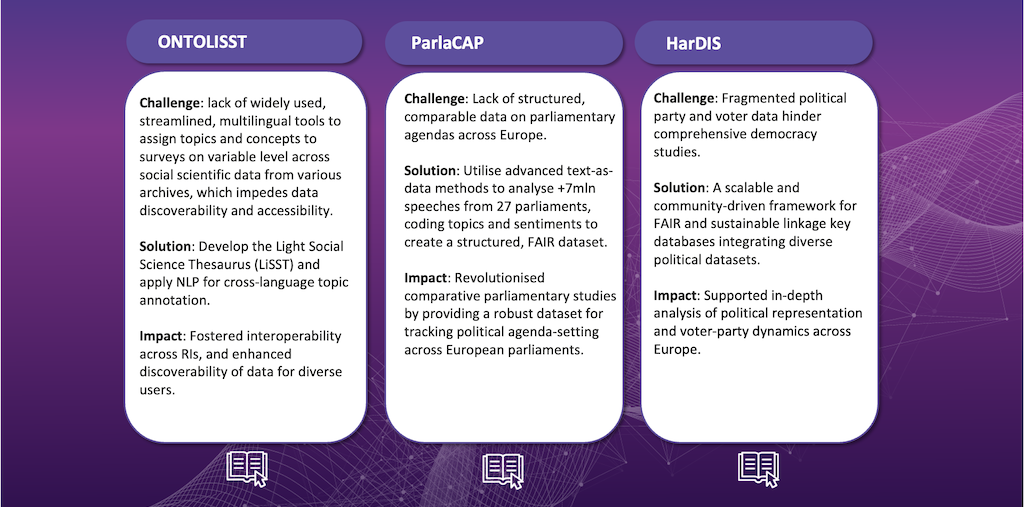
Below is an overview of the projects working to make improvements primarily in this area:
Together, these efforts affirm OSCARS’ role as a catalyst for improving the semantic and structural clarity of research data - an essential step toward a more FAIR, federated, and inclusive Open Science ecosystem.


Browse all OSCARS funded projects here
Useful resources:
Policy & Strategic Frameworks
- EOSC SRIA 1.3 (Strategic Research and Innovation Agenda), Version 1.3 (2024)
Outlines EOSC’s roadmap, including semantic interoperability, FAIR data workflows, and incentive mechanisms. - European Commission: Turning FAIR into Reality (2018)
Foundational report defining FAIR principles and the need for annotation and metadata standards.
FAIR Principles & Annotation Guidance
- GO FAIR Initiative – FAIR Principles
Practical guidance on FAIR data stewardship, annotation practices, and FAIR Implementation Profiles. - FAIR Implementation Profiles (FIPs) Wizard
Tool co-developed with GO FAIR, used by ENVRI and other RIs to structure metadata and annotation practices. - FAIRsharing.org
A registry of metadata standards, databases, and data policies—very useful for selecting annotation standards.
Additional Annotation & Metadata Resources
- FAIR Cookbook (ELIXIR & FAIRplus)
Hands-on recipes for FAIRifying data, including annotation workflows in life sciences. - FAIR Digital Object (FDO) Forum - Framework for structuring annotated digital objects with persistent identifiers.
Authors
Nicoletta Carboni (CERIC-ERIC), Romain David (ERINHA), Franciska de Jong (Utrecht University), Jonathan Ewbank (ERINHA), Darja Fišer (CLARIN ERIC), Björn Grüning (University of Freiburg), Justine Thomas (CNRS-LAPP), Friederike Schmidt-Tremmel (Trust-IT).
DOI
http://doi.org/10.5281/zenodo.17224943
References
[1] The FAIR Implementation Profile (FIP) Wizard is a tool, built on the Data Stewardship Wizard software, that allows research communities to document their specific choices for implementing the FAIR Guiding Principles.